AutoDock Vina
This site was built for the legacy version of AutoDock Vina, v1.1.2 (last revision: May 2011). It remains open for information purposes. Most of the methods and protocols for protein-ligand docking have been re-implemented with improvements in the current docking engines:
AutoDock Vina v1.2.x (2021–present): https://github.com/ccsb-scripps/AutoDock-Vina
AutoDock-GPU (2021–present): https://github.com/ccsb-scripps/AutoDock-GPU
For the latest developments and information, please visit the linked project Github pages or our new navigation & resource site at https://rsd3.scripps.edu/.
 AutoDock Vina is an open-source program for doing molecular docking. It was originally designed and implemented by Dr. Oleg Trott in the Molecular Graphics Lab (now CCSB) at The Scripps Research Institute.
AutoDock Vina is an open-source program for doing molecular docking. It was originally designed and implemented by Dr. Oleg Trott in the Molecular Graphics Lab (now CCSB) at The Scripps Research Institute.
The latest version is available here.
AutoDock Vina is one of the docking engines of the AutoDock Suite.
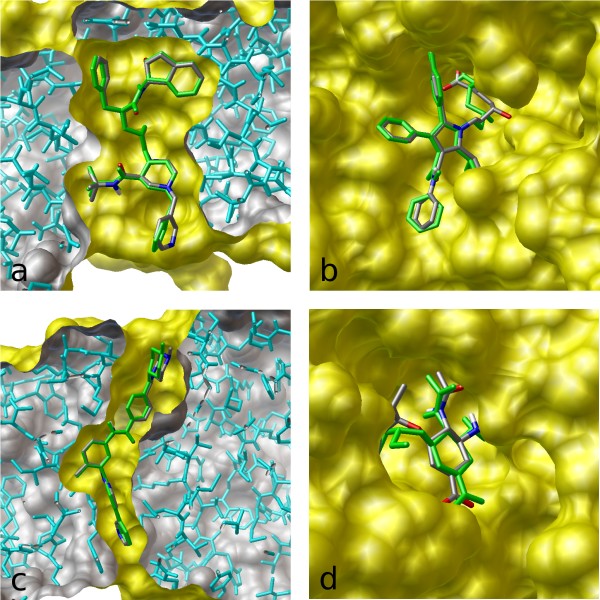
The image on the left illustrates the results of flexible docking (green) superimposed on the crystal structures of (a) indinavir, (b) atorvastatin, (c) imatinib, and (d) oseltamivir bound to their respective targets.
Publications
If you used AutoDock Vina in your work, please cite:
Features
| Accuracy
AutoDock Vina significantly improves the average accuracy of the binding mode predictions compared to AutoDock 4, judging by our tests on the training set used in AutoDock 4 development.[*] Additionally and independently, AutoDock Vina has been tested against a virtual screening benchmark called the Directory of Useful Decoys by the Watowich group, and was found to be “a strong competitor against the other programs, and at the top of the pack in many cases”. It should be noted that all six of the other docking programs, to which it was compared, are distributed commercially. AutoDock Tools Compatibility For its input and output, Vina uses the same PDBQT molecular structure file format used by AutoDock. PDBQT files can be generated (interactively or in batch mode) and viewed using MGLTools. Other files, such as the AutoDock and AutoGrid parameter files (GPF, DPF) and grid map files are not needed. |
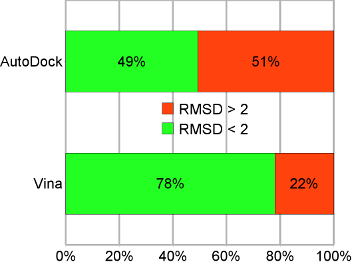 Binding mode prediction accuracy on the test set. “AutoDock” refers to AutoDock 4, and “Vina” to AutoDock Vina 1. |
Ease of Use
Vina’s design philosophy is not to require the user to understand its implementation details, tweak obscure search parameters, cluster results or know advanced algebra (quaternions). All that is required is the structures of the molecules being docked and the specification of the search space including the binding site. Calculating grid maps and assigning atom charges is not needed. The usage summary can be printed with “vina --help“. The summary automatically remains in sync with the possible usage scenarios.
Implementation Quality
- By design, the results should not have a statistical bias related to the conformation of the input structure.
- Attention is paid to checking the syntactic correctness of the input and reporting errors to the user in a lucid manner.
- The invariance of the covalent bond lengths is automatically verified in the output structures.
- Vina avoids imposing artificial restrictions, such as the number of atoms in the input, the number of torsions, the size of the search space, the exhaustiveness of the search, etc.
Flexible Side Chains
Like in AutoDock 4, some receptor side chains can be chosen to be treated as flexible during docking.
| Speed
AutoDock Vina tends to be faster than AutoDock 4 by orders of magnitude.[*] Multiple CPUs/Cores Additionally, Vina can take advantage of multiple CPUs or CPU cores on your system to significantly shorten its running time. World Community Grid Qualified projects can run AutoDock Vina calculations for free on the massively parallel World Community Grid. Existing projects using AutoDock Vina there include those targeting AIDS, Malaria, Leishmaniasis and Schistosomiasis. Some of these projects average over 50 years worth of computation per day. |
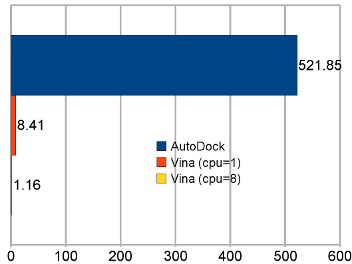 Average time per receptor-ligand pair on the test set. “AutoDock” refers to AutoDock 4, and “Vina” to AutoDock Vina 1. |
License
AutoDock Vina is released under a very permissive Apache license, with few restrictions on commercial or non-commercial use, or on the derivative works. The text of the license can be found here.
Video Tutorial
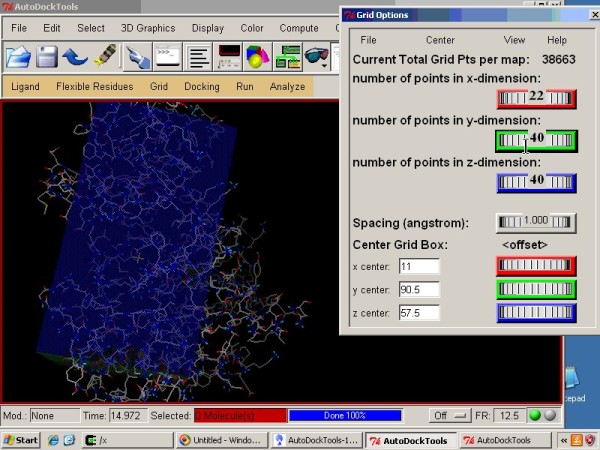 |
This video tutorial demonstrates molecular docking of imatinib using Vina with AutoDock Tools and PyMOL |